Jupyter Notebooks¶
Jupyter notebooks provide a way to run code in an interactive environment. While most prominently used for Python, Jupyter supports a range of languages, such as Julia and R.
Choosing an Approach¶
There are multiple ways to run Jupyter notebooks on the computing clusters available through ORCD. The route you choose depends on your needs and level of familiarity with high performance computing environments.
Web Portal¶
The most straightforward way to run a Jupyter notebook on one of our computing clusters is to use the cluster's web portal. While this route is the easiest to set up, it can be limiting if you want more control over your environment or the resources allocated to your notebook.
-
Link to web portal: https://engaging-ood.mit.edu/
-
Select "Interactive Apps" --> "Jupyter Notebook"
-
Follow the on-screen instructions to start a session. You are able to use a custom Conda environment provided it has
jupyterlabinstalled. -
If you'd like to run Julia, enter the name of the Julia module you're using (e.g.,
julia/1.8.5). Note that you need to haveIJuliainstalled in your environment for this version of Julia. -
If you'd like to run R, enter the name of your custom Conda environment that has
r-irkernelinstalled. -
When the session is ready, click "Connect to Jupyter." From here you can create a Jupyter notebook and select the language you would like to use.
Note
The Engaging web portal is currently running on CentOS 7, which has a different set of modules from Rocky 8 nodes. If you would like to run a Jupyter notebook on Rocky 8, you will need to follow either the VS Code or port forwarding method.
VS Code¶
First, follow these instructions to set up VS Code to run on a compute node.
Open a Jupyter notebook and click the top right button to select a kernel. You
can select "Python Environments" for any Conda environments or "Jupyter Kernel"
to find Julia or R environments. If you have installed R with Conda, you can
find your Conda environment under "Jupyter Kernel." jupyterlab must be
installed to your Conda environment.
Port Forwarding¶
Port forwarding offers the most flexibility in setting up your Jupyter notebook but the setup is slightly more involved. With port forwarding, the rendering is handled through your internet browser while computation is done on the cluster. This method is more lightweight than VS Code and can be more reliable.
Port forwarding consists of running the notebook on a compute node and then accessing the notebook on your local machine by SSH tunnelling through a login node.
First, request a compute node with the resources you'll need for your Jupyter session (here we are requesting 1 node with 4 CPU cores):
Note
See Requesting Resources for more information.
Make a note of the node that your job is running on by running hostname from
the command line.
Even if you are using a different language with Jupyter, Jupyter is tightly
linked to Python, so you will need to use a Conda environment with
jupyterlab installed:
Now, we can run the notebook. To be able to access it on our local machine, we need to add a few arguments:
port=$(python -c 'import socket; s=socket.socket(); s.bind(("", 0)); print(s.getsockname()[1]); s.close()')
jupyter-lab --ip=0.0.0.0 --port=$port
The port can be any number between 1024 and 65535, and the first command above ensures getting an open port.
When you run the notebook, the output will contain a link with a token that allows you to access the notebook, which will look like the following:
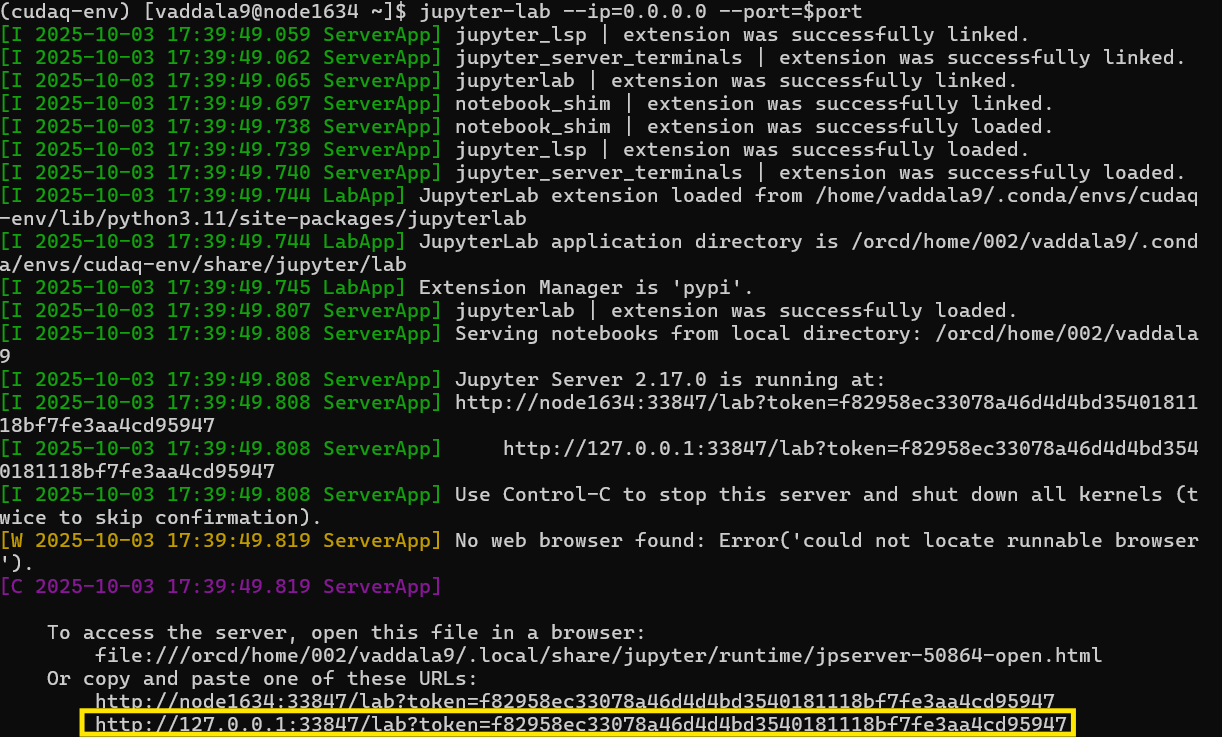
Make sure to select the second URL that is provided as outlined in yellow above. We cannot use this link right away because that node is not available from our local machine. Through "tunneling," however, we can access this node through a login node, which is accessble from our local machine.
In a second terminal window on your local machine, set up an SSH tunnel to your Jupyter notebook that's running on the compute node, filling in the node name, port number, and username as necessary. We will keep the local port and the remote port the same for simplicity.
Now you can access Jupyter in an internet browser using the link we received above.
Now you can open a Jupyter notebook and select your kernel from the top right corner. The Python environment is the same environment you used to run the notebook.
Language-Specific Instructions¶
Julia¶
You will need to add the IJulia package to your environment for Jupyter to
recognize the Julia kernel. You can do so from the command line:
Unlike R, Julia environments are separate from Conda. However, if the IJulia
package is installed, then the Julia kernel should be visible regardless of the
Conda environment from which you run your Jupyter notebook.
See our Julia documentation for more information.
R¶
To run R in a Jupyter notebook, you need to create a Conda environment with
both r-irkernel and jupyterlab installed:
module load miniforge
conda create -n r_jupyter_env jupyterlab r-irkernel
conda activate r_jupyter_env
Most R packages are available through Conda, so feel free to install other packages you need to this environment.
See our R documentation for more information.
Python¶
To run Python Jupyter notebooks, install jupyterlab to whatever Conda
environment that contains the packages you need.
See our Python documentation for more information.
FAQs¶
How do I run a Jupyter notebook with a GPU?
The cluster web portals offer an option to allocate a GPU to your Jupyter session. If you would like to use a different partition, however, then follow the instructions for VS Code or port forwarding and request a GPU in your Slurm job. See our documentation on requesting resources for more information.
Jupyter does not recognize the kernel for my environment. What do I do?
First, make sure you have r-irkernel installed if you're using R, IJulia
installed (and built) if you're using Julia, and jupyterlab installed to
your Conda environment.
On VS Code, you may need to specify the path to the conda binary of the Conda
installation you're using. This can be done by editing the "Python: Conda Path"
setting. For example, if you're using the miniforge/24.3.0-0 module on
Engaging, the path would be:
To see all kernels that Jupyter recognizes, activate a Conda environment with
jupyterlab installed and run jupyter kernelspec list.
I tried to install jupyterlab to my Conda environment, but the installation
failed. How can I run a Jupyter notebook with the dependencies I need?
It's best to install the packages you need when you create a Conda environment rather than one-by-one after the environemnt has been created. This will make Conda more likely to solve your environment succesfully. For example:
See Conda Environments for more information.